Experimenting with the tripalv3 JSON+LD API
Introduction
In this post I'm experimenting with querying the JSON-LD (LD stands for Linked Data) API of my Tripal3 genome database for Pseudomonas fluorescens SBW25. A little bit of background: Tripal is a content management system (CMS) for genomic data, based on the Drupal CMS framework. It facilitates setting up customized genomic database webservers to publish genome assemblies, annotation tracks, experimental and modeling data for one or several organisms. The current stable release is Tripal-v3. Tripal implements the Chado database schemefor biological (genomic) databases Popular examples for Tripal instances are the Rice Genome Hub, or the Kiwifruit Genome Database, more examples can be found on the Tripal website. What sets Tripal aside compared to other webserver frameworks with similar objectives (e.g. machado, the UCSC Genome Browser, or the Ensembl Genome Browser), is that all data in the Tripal database is accessible through a JSON API. Moreover, the JSON response adheres to the JSON-LD standard issued by the World Wide Web Consortium W3C. This feature makes Tripal (in principle) compatible with Semantic Web applications and eases the connectivity of Linked Data services such as public SPARQL endpoints and Tripal sites or between Tripal sites.
Accessing Tripal data as JSON objects
The entry point to all Tripal data formatted in JSON is the canonical webservice address http://<tripal-server-url>/web-services/content/v0.1/. Here "<tripal-server-url> stands for the homepage address of any Tripal instance. In my case, it would be pflu.evolbio.mpg.de. The last element of the webservices URL indicates the API version which is currently 0.1.
Below is a screenshot of our Tripal instances JSON webservice URL accessed in a web browser.
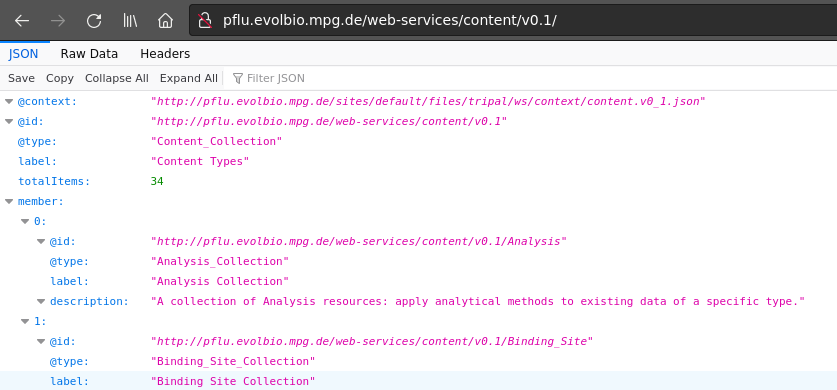
The first entry loads a web resource, the context. The context is a kind of glossary that defines web prefixes for the common parts of frequently used IRIs in the document (more details about context here). Next come the id, type, and label of the document, followed by the number of items in the document. The member list containes all the top level entities, in particular the classes that correspond to the various feature types in the genome. Following the link of a given feature type opens the corresponding (JSON-LD) document as shown here for the CDS Collection:

Again, the document's context, id, type, and label are given, followed by a count, a view section defining parameters for web display and finally the member list containing all CDS features in the database along with their respective id, type, label and the URL where all available information (annotations, cross references, sequence coordinates, …) is displayed.
JSON-LD and SPARQL
I'm working on a data integration task where I want to use Linked Data technology to connect multiple data sources with each other. Our Tripal site is one of these data sources. For quite some time I've been exploring ways how to access the data in the relational database underlying the Tripal site using SPARQL. My first choice was to reformat the GFF3 annotation file to RDF and serve that through a fuseki server. It works but has the obvious drawback of data duplication because the same GFF3 file also goes into the Tripal site to populate its DB. Next try was to use a mapping to tranlate SPARQL queries into SQL queries which would then be run on the Tripal DB and results would be backtranslated into RDF. I presented the results of that work at the last Bioontologies conference at ISMB2022. We used the Ontop-VKG solution to bootstrap the mapping and ontology and to serve the resulting virtual knowledge graph in a SPARQL endpoint. Example queries can be found in this blog post. The drawback of this approach is that Ontop-VKG (as well as other mapping/VKG solutions to my knowledge) do not support federated queries, in my opinion the major stronghold of the entire RDF and SPARQL technology. Secondly, aligning the bootstrapped ontologies with the ontologies that are used in the Tripal database and the chado database schema is a formidable task in itself.
I've had it in the back of my head since starting out on this project that it should be possible to run a SPARQL query on the Tripal database directly through the JSON API (even posted a github issue to this effect). Recently, I came across Bob DuCharme's blog post about Exploring JSON-LD and I've now started experimenting in a similar manner with the JSON-LD data of our Tripal site. Whereas Bob "wrote a script to pull the JSON-LD" (I believe using some sort of webscraping), my case is much more comfortable since there is a dedicated JSON API.
SPARQL queries against our Tripal site
Let's start with a simple example, exploring the top level JSON API entry point. One thing to be aware of is that the Tripal JSON API is not a SPARQL endpoint, hence SPARQL clients such as python's SPARQLWrapper, jupyter's sparqlkernel, or org's sparql-mode cannot be used (the latter is rather sad since I'm writing this post in org). Here, I'm using the arq client that comes with the Apache Jena package.
My query reads
# sparql query file sop.rq
select * where {
?s ?p ?o .
}
limit 10Running the following arq command (my Apache Jena installation is in a non-standard location, hence I give the complete path to arq):
/opt/apache-jena/bin/arq \
--data http://pflu.evolbio.mpg.de/web-services/content/v0.1/ \
--query sop.rq------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | s | p | o | =============================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================== | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Genetic_Marker> | <http://www.w3.org/2000/01/rdf-schema#comment> | "A collection of Genetic Marker resources: a measurable sequence feature that varies within a population. [SO:db]" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Genetic_Marker> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://pflu.evolbio.mpg.de/web-services/doc/v0.1#Genetic_Marker_Collection> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Genetic_Marker> | <http://www.w3.org/2000/01/rdf-schema#label> | "Genetic Marker Collection" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Pseudogene> | <http://www.w3.org/2000/01/rdf-schema#comment> | "A collection of Pseudogene resources: a sequence that closely resembles a known functional gene, at another locus within a genome, that is non-functional as a consequence of (usually several) mutations that prevent either its transcription or translation (or both). In general, pseudogenes result from either reverse transcription of a transcript of their \\\"normal\\\" paralog (SO:0000043) (in which case the pseudogene typically lacks introns and includes a poly(A) tail) or from recombination (SO:0000044) (in which case the pseudogene is typically a tandem duplication of its \\\"normal\\\" paralog). [http://www.ucl.ac.uk/~ucbhjow/b241/glossary.html]" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Pseudogene> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://pflu.evolbio.mpg.de/web-services/doc/v0.1#Pseudogene_Collection> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Pseudogene> | <http://www.w3.org/2000/01/rdf-schema#label> | "Pseudogene Collection" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Heritable_Phenotypic_Marker> | <http://www.w3.org/2000/01/rdf-schema#comment> | "A collection of Heritable Phenotypic Marker resources: a biological_region characterized as a single heritable trait in a phenotype screen. The heritable phenotype may be mapped to a chromosome but generally has not been characterized to a specific gene locus. [JAX:hdene]" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Heritable_Phenotypic_Marker> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://pflu.evolbio.mpg.de/web-services/doc/v0.1#Heritable_Phenotypic_Marker_Collection> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Heritable_Phenotypic_Marker> | <http://www.w3.org/2000/01/rdf-schema#label> | "Heritable Phenotypic Marker Collection" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Phylogenetic_Tree> | <http://www.w3.org/2000/01/rdf-schema#comment> | "A collection of Phylogenetic Tree resources: the raw data (not just an image) from which a phylogenetic tree is directly generated or plotted, such as topology, lengths (in time or in expected amounts of variance) and a confidence interval for each length." | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
As expected, the query returns the first ten (randomly chosen) entries of the JSON API entry point. Let's run the same query on the CDS entry point http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS :
/opt/apache-jena/bin/arq \
--data http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS \
--query sop.rq----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | s | p | o | ================================================================================================================================================================================================================= | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11856> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11856> | <https://schema.org/ItemPage> | "http://pflu.evolbio.mpg.de/bio_data/11856" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11856> | <http://www.w3.org/2000/01/rdf-schema#label> | "CDS:PFLU_0012-0" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11862> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11862> | <https://schema.org/ItemPage> | "http://pflu.evolbio.mpg.de/bio_data/11862" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11862> | <http://www.w3.org/2000/01/rdf-schema#label> | "CDS:PFLU_0018-0" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS?page=1&limit=25> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/PartialCollectionView> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11868> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11868> | <https://schema.org/ItemPage> | "http://pflu.evolbio.mpg.de/bio_data/11868" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11868> | <http://www.w3.org/2000/01/rdf-schema#label> | "CDS:PFLU_0024-0" | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Which returns the first 10 CDS records. So far so good so expected. These are two typical queries to explore the dataset. Now imagine the following scenario: I run the first query (on the top level entry point) and find that there is a CDS collection. I want to explore the CDS collection by brachiating through the graph. I have to write my query such that it uses the subject IRI of the CDS collection http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS in a FROM clause for a subsequent (sub)query. Unfortunately, using "FROM" in a subquery does not seem to be possible as discussed in this StackOverflow entry. Running a federated query is also not possible, again because the JSON API is not a proper SPARQL endpoint.
The following query:
# sparql query file cds_from_entrypoint.rq
select *
from <http://pflu.evolbio.mpg.de/web-services/content/v0.1>
from <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS>
where {
?cds_collection a <http://pflu.evolbio.mpg.de/web-services/doc/v0.1#CDS_Collection> .
?cds_collection <http://www.w3.org/ns/hydra/core#member> ?cds .
?cds <https://schema.org/ItemPage> ?item_page .
?cds a ?type .
}
limit 5works though (note that I'm now passing the data source in a "from" clause in the query file):
/opt/apache-jena/bin/arq --query cds_from_entrypoint.rq------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | cds_collection | cds | item_page | type | ======================================================================================================================================================================================================================================================= | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS> | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11858> | "http://pflu.evolbio.mpg.de/bio_data/11858" | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS> | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11863> | "http://pflu.evolbio.mpg.de/bio_data/11863" | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS> | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11852> | "http://pflu.evolbio.mpg.de/bio_data/11852" | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS> | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11857> | "http://pflu.evolbio.mpg.de/bio_data/11857" | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS> | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11846> | "http://pflu.evolbio.mpg.de/bio_data/11846" | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Which gives me the item webpage for every feature in the CDS Collection. What's nice is that I don't have to rewrite the arq command, but the obvious drawback is that I need to hardcode the CDS entry point into the query file (2nd "from" above).
Querying CDS properties
Now let's dive a bit deeper and try to query some feature properties of our favorite CDS PFLU_0006. I first have to retrieve the entries data entrypoint by querying the JSON document holding the CDS Collection:
# sparql query file cds_properties.rq
select *
from <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS>
where {
?s a <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> .
?s <http://www.w3.org/2000/01/rdf-schema#label> "CDS:PFLU_0006-0" .
?s <http://schema.org/ItemPage> ?item_page .
}which, when run through arq, returns
/opt/apache-jena/bin/arq --query cds_properties.rq------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | s | p | o | ================================================================================================================================================================================================ | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11850> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11850> | <http://www.w3.org/2000/01/rdf-schema#label> | "CDS:PFLU_0006-0" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/11850> | <https://schema.org/ItemPage> | "http://pflu.evolbio.mpg.de/bio_data/11850" | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
As expected, I get the item page (for web access) and the entries Linked Data URI for programmatic access. However, modifying the above query to get hold of another CDS,
# sparql query file cds_properties.rq
select *
from <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS>
where {
?s a <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> .
?s <http://www.w3.org/2000/01/rdf-schema#label> "CDS:PFLU_0408-0" .
?s <http://schema.org/ItemPage> ?item_page .
}the same query
/opt/apache-jena/bin/arq --query cds_properties.rq------------- | s | p | o | ============= -------------
returns an empty result. What happened? Pagination kicked in: not the entire JSON document is parsed but only the top 25 entries as per pagination (view) settings in the JSON document. The JSON API accepts the parameters "page" and "limit" to control pagination. "page" sets the page number to be returned and "limit" the number of items per page. "page" defaults to 1, hence to retrieve all items, it would be sufficient to set only the "limit" parameter to a value larger or equal to the number of items. Be aware that the response time scales with the number of elements per page, the default 25 is a good compromise between responsiveness and number of items. Updating the above query as
# sparql query file cds_properties.rq
select *
from <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS?page=39&limit=10>
where {
?s a <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> .
?s <http://www.w3.org/2000/01/rdf-schema#label> "CDS:PFLU_0408-0" .
?s <http://schema.org/ItemPage> ?item_page .
}and run this through arq:
/opt/apache-jena/bin/arq --query cds_properties.rq------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | s | p | o | ================================================================================================================================================================================================ | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://www.w3.org/2000/01/rdf-schema#label> | "CDS:PFLU_0408-0" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <https://schema.org/ItemPage> | "http://pflu.evolbio.mpg.de/bio_data/12234" | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
gives the expected output for the CDS at locus PFLU_0408.
Now lets extend the last query to gather all properties of CDS:PFLU_0408-0, such as annotations, genome coordinates etc. From the previous query, we have the subject URI of our target CDS, which we now include in our query in a "from" clause:
# sparql query file cds_properties.rq
select *
from <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS?page=39&limit=10>
from <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234>
where {
?s a <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> .
?s <http://www.w3.org/2000/01/rdf-schema#label> "CDS:PFLU_0408-0" .
?s ?p ?o .
} /opt/apache-jena/bin/arq --query cds_properties.rq------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | s | p | o | ============================================================================================================================================================================================================================================================================================================================================================================== | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/frame> | "0" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://edamontology.org/data_2044> | "http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234/Sequence" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <https://schema.org/name> | "CDS:PFLU_0408-0" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/similarity> | "fasta; with=UniProt:Q51352_PSEAE (EMBL:AE004917); Pseudomonas aeruginosa.; pilN; Type 4 fimbrial biogenesis protein PilN.; length=198; id 42.135%; ungapped id 42.857%; E()=2e-18; 178 aa overlap; query 4-178; subject 3-180" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://edamontology.org/data_0842> | "CDS:PFLU_0408-0" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/codon_start> | "1" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/contact> | "http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234/contact" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://www.w3.org/2000/01/rdf-schema#type> | "CDS" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/inference> | "Predicted" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/protein_id> | "CAY46685.1" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://purl.obolibrary.org/obo/OBI_0100026> | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/Organism/1> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://purl.obolibrary.org/obo/SBO_0000374> | "http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234/relationship" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://edamontology.org/data_2091> | "" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <https://schema.org/publication> | "http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234/publication" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/seqid> | "MPBAS00001" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://edamontology.org/data_2190> | "d41d8cd98f00b204e9800998ecf8427e" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/product> | "Putative fimbriae biogenesis-related protein" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://purl.obolibrary.org/obo/OGI_0000021> | "http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234/location+on+map" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/timeaccessioned> | "2022-02-24 22:06:44" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/uniprot_annotation_score> | "1 out of 5" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://purl.obolibrary.org/obo/SBO_0000554> | "http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234/database+cross+reference" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> | <http://www.sequenceontology.org/browser/current_svn/term/SO:0000316> | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/features> | "Coiled coil (1); Transmembrane (1)" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/timelastmodified> | "2022-02-24 22:06:44" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/uniprot_review_status> | "unreviewed" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/locus_tag> | "PFLU_0408" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/is_analysis> | false | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://semanticscience.org/resource/SIO_001166> | "http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234/annotation" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://edamontology.org/data_2012> | "http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234/Sequence+coordinates" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://www.w3.org/2000/01/rdf-schema#label> | "CDS:PFLU_0408-0" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <https://schema.org/ItemPage> | "http://pflu.evolbio.mpg.de/bio_data/12234" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/gene> | "PFLU_0408" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/confidence_level> | "3" | | <http://pflu.evolbio.mpg.de/web-services/content/v0.1/CDS/12234> | <http://pflu.evolbio.mpg.de/cv/lookup/local/is_obsolete> | false | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
which now lists all the information and links for CDS:PFLU_0408-0.
In conclusion, we successfully queried the database underlying our Tripal genome website using SPARQL. For each query, we have to hardcode the URI of the targeted entity, which can be obtained from a query on the parent JSON object that links to the target object. What we cannot do is to query the subject URI and retrieve the subject's triples in a single query. For example, in the above, we could remove the first "from" clause and still get the same information.
In other words, the tripal JSON API is a collection of linked named graphs, each living in its own JSON document. In order to support SPARQL queries that brachiate through the hierarchy, we would have to construct a RDF tree that includes all nodes. This could be achieved through script that, starting from the JSON API entry point, recursively iterates through all "members" and constructs all the triples (quads) it encounters. These quads could then be stored in a triplestore, e.g. jena-fuseki or virtuoso to allow better responsiveness also for complex queries. I'll try that in a forthcoming post.